You, Stoomboot, and a thesis as a present
Nikhef Computing Course, Tuesday 2022-11-22
Dennis van Dok
Learning goals
- How to submit a batch job
- Batch system queues and available resources
- CVMFS, containers and conda
- Write a batch job description file
- Batch processing
- Know how to write data to dcache using data transfer protocols such as xrootd and webdav
- Understand the pitfalls for scaling up batch jobs
Organising your computer work
- Separate code from data.
- Understand where each of these things go
- Setting up your computational environment
- for reproducible results
- for scaling up
- Sharing and publishing your results
Overview of computer resources
All resources are shared among our users.
There is no automated restriction or intervention, we don't want to throw up barriers.
Please apply constraint, and observe the social contract in civil discourse among colleagues:
- don't assume bad intent
- inform, but don't blame
- involve experts to work out a solution
Interactive nodes
We have two (soon: three) heavy-duty compute nodes with 32 cores each for
- compiling software
- running tests
- short analysis jobs
They are called stbc-i{1,2,3} and can be reached by ssh.
GPU nodes
We have a number of GPU nodes; for each type we have an interactive node that you can log on to for running tests.
You can't reserve these nodes, so don't abuse them for long runs.
We also have identical batch nodes so production runs can be scheduled and guaranteed exclusive use of the node.
- The stoomboot documentation page on GPU nodes
- https://wiki.nikhef.nl/ct/Stoomboot_cluster#GPU_Nodes
Batch processing

What is batch processing
A.k.a. the original computing.
Batch systems run programs in a non-interactive way. The user has to specify up front what the parameters to the program are. The program is submitted to the head node and queued for running at a later time.
Stoomboot

Torque
The stoomboot batch system is running Torque with Maui for scheduling. This is an old but proven piece of technology.
Jobs are presented to the head node from any one of the interactive nodes or even login or desktop nodes.
Scheduling of batch jobs
This is based on several factors, such as resources available, resources requested, relative priority, resource use in the last 24 hours and others. It is therefore hard to predict what the ordering of jobs is going to be.
The job is run on one of the worker nodes, likely concurrently with other user's jobs. Since these are multi-core machines, a job usually has full use of the computing power of a single core.
Worker nodes
The stoomboot batch farm has around 2k cores. There are two main types of nodes
| node type | CPU | number | total cores | used by |
|---|---|---|---|---|
| sate | AMD EPYC 7551P 32-Core Processor | 25 | 800 | all |
| knek | AMD EPYC 7702P 64-Core Processor | 18 | 1152 | smefit* |
*The knek nodes can be used opportunistically by jobs in the short queue.
Resource limitations
Worker nodes are dimensioned to have 8GB per core. You can request more memory for your job if needed and this will influence scheduling as the scheduler will have to fit all the memory requests of jobs into main memory.
A batch job runs on a single core, but multiple cores can be requested. This will also have an impact on the scheduling.
Submitting a job
- Simple submission
qsub my_job.sh
- Adding parameters like resource requests
qsub -q short -l walltime=2:00:00 my_job.sh
- More memory, more cores, or more time
qsub -l nodes=1:ppn=4 -l pvmem=16gb -l walltime=72:00:00 job.sh
Resource requests can also go into the job script
myjob.sh:
#!/bin/sh
#PBS -q long
#PBS -l mem=192gb
#PBS -l nodes=1:ppn=24
#PBS -l cput=17280:00:00
#PBS -l walltime=720:00:00
# now burn some CPU time
${HOME}/myproject/bin/analyse.sh
Inspecting the state of your jobs
- Return a list of all jobs
qstat
- Only my jobs
qstat -u $USER
- Everything including array jobs and worker nodes
qstat -ant1
- Kill all my queued jobs
qselect -s Q -u $USER | xargs qdel
State of jobs (2)
Req'd Req'd Elap
Job ID Username .. Memory Time S Time
----------------------- -------- .. ------ --------- - ---------
12223328.burrell.nikhe ...svd .. -- 24:00:00 R 00:01:52
The state column (S) indicates where your jobs is at:
| Q | queued |
| R | running |
| E | exiting* |
| C | completed |
| H | Held* |
| W | Waiting* |
*These are transitional; if you notice a job in this state for longer than a minute let us know!
Queues
$ qstat -Q Queue Max Tot Ena Str Que Run Hld Wat Trn Ext T Cpt gpu7 0 0 yes yes 0 0 0 0 0 0 E 0 gpu 0 0 yes yes 0 0 0 0 0 0 R 0 express 0 0 yes yes 0 0 0 0 0 0 R 0 gpu-amd 0 0 yes yes 0 0 0 0 0 0 E 0 gpu-nv 0 0 yes yes 0 0 0 0 0 0 E 0 smefit 0 1060 yes yes 825 235 0 0 0 0 E 0 short 0 0 yes yes 0 0 0 0 0 0 R 0 long7 0 67 yes yes 0 24 0 0 0 0 E 43 generic 0 0 yes yes 0 0 0 0 0 0 R 0 long 0 0 yes yes 0 0 0 0 0 0 R 0 multicore 0 0 yes yes 0 0 0 0 0 0 R 0 generic7 0 984 yes yes 705 278 0 0 0 0 E 1 short7 0 4443 yes yes 4162 172 0 0 0 0 E 109 express7 0 0 yes yes 0 0 0 0 0 0 E 0
The E queues are execution queues; you can submit to one of routing queues (R) and the system will send them to an appropriate E queue.
Queues
- express
- At most two jobs per user; at most 10 minutes of run time. This is for really short tests but has the most likely chance of getting to run within minutes of submission
- short
- At most 4 hours of run time and 8 GB of memory per core. Preferred for production use if the jobs fit. Best for a couple of runs per day.
- generic
- At most 24 hours of run time and 16 GB of memory per core. The default queue.
- long
- Up to 96 hours and 16 GB RAM. We limit the total amount of long jobs to have decent turn around time of short job
- multicore
- Obsolete, any queue can be used with multiple cores.
- smefit
- Exclusively for the smefit group, will use the knekel nodes.
GPU queues
These queues are for jobs that make use of the GPU cluster, so not for general CPU use.
- gpu
- matches any GPU node. Unless your job is GPU brand agnostic, probably not a good idea to use
- gpu-amd
- matches any of the AMD MI50 nodes
- gpu-nv
- matches any of the NVIDIA GPU nodes
Matching on specific GPU cards
Given the variety of GPUs in our systems, you may want to direct your work to a specific type. That can be done by matching on node properties.
qsub -l 'nodes=1:mi50' -q gpu job.sh
| type | brand | nodes | total gpus | property |
|---|---|---|---|---|
| GeForce GTX 1080 | NVIDIA | 2 | 2 | gtx1080 |
| Tesla V100 | NVIDIA | 1 | 2 | v100 |
| Radeon Instinct MI50 | AMD | 6 | 12 | mi50 |
More info
Documentation available on-line:
https://support.adaptivecomputing.com/torque-resource-manager-documentation/
See the EOL versions→4.2.x
Software management
CernVM-FS
The CernVM file system (CVMFS) is one of the greatest successes to come out of CERN IT since the world wide web. (IMHO)
Today it is used worldwide by various science communities for distributing their software in a highly scalable, robust and high-performance manner.
CVMFS mechanism
- Original data is maintained on a Stratum-0 server
- When an update is published everything is broken into unique objects (so we have de-duplication)
- Everything is mirrored onto several Stratum-1 servers over HTTP
- Worker nodes that want to retrieve files from the repository request the object via HTTP and a local squid proxy
- Requests are cached locally and at the squids so repeated retrievals will not do another retrieval
Your project likely already uses CVMFS in some capacity, so you should check with them.
CVMFS hierarchy
- CVMFS Documentation
- https://cvmfs.readthedocs.io/en/stable/
Finding software
Many common software packages are made available for various platforms by the CERN SFT team.
E.g. numpy:
https://lcginfo.cern.ch/pkgver/numpy
- Check the versions; then check the releases.
Find the same under
/cvmfs/sft.cern.ch/lcg/releases/LCG_<release>/numpy/<version>
Then, under the correct subdirectory for your platform and compiler combination:
$ source /cvmfs/sft.cern.ch/lcg/releases/LCG_102/numpy/1.22.3/⮐
x86_64-centos7-gcc8-opt/numpy-env.sh
$ python
Python 3.9.12 (main, Jun 7 2022, 16:10:25)
[GCC 8.3.0] on linux
Type "help", "copyright", "credits" or "license" for more information.
>>> import numpy
>>> numpy
<module 'numpy' from '/cvmfs/sft.cern.ch/lcg/releases/LCG_102/numpy/1.22.3/⮐
x86_64-centos7-gcc8-opt/lib/python3.9/site-packages/numpy/__init__.py'>
Controlling the software environment
For reproducible science and you overall sanity it's vital to work within a well-defined environment. Being in control over versions of the many software libraries that you employ can be difficult if you rely on whatever the platform provides.
There are a couple of popular approaches here, but the choice may not be yours to make. You will likely go with what is already out there in your particular field or group.
Python virtual environments
Python brings the concept of virtual environments. Using this changes the environment variables for finding binaries and packages to isolate you from the setup of the base system. Maintaining separate virtual environments is possible as well.
Conda
A closely related alternative is to use conda. This is more of a generic distribution system. Conda environments can get very large very quickly, so don't put them in your home directory.
conda config --append envs_dirs /project/myfirstproject/conda/envs
conda create --prefix /project/myfirstproject/conda/envs/shared_python38⮐
python=3.8
conda activate shared_python38
conda install scipy --channel conda-forge
Virtualenv or Conda?
"It depends":
Containers
Another approach is to pack everything and the kitchen sink in a container.
- Running software in a container is conceptually like running on a completely different machine
- The only thing that the container shares with its host operating system is the running kernel.
- Everything else, the file system tree, the process tree, the network connections and the system users can be isolated.
Docker
Software like Docker made this very popular and there is an open marketplace of basic container images for every thinkable application on Dockerhub.
Modern Linux kernels offer the capability of letting unprivileged users run unpacked containers. An unpacked container is basically a file system tree, so it can be copied around like an ordinary directory.
Apptainer and CVMFS
This is extremely powerful in combination with CVMFS!
- Unpacked containers are stored on CVMFS
- When used, individual files are retrieved instead of the entire image
- better caching!
- de-duplication! Have multiple containers all alike and store only the changes!
- Works anywhere!
Converting other container formats
Be sure to set the APPTAINER_CACHEDIR environment to somewhere
outside your home directory; this could get large.
mkdir /tmp/mydir && cd /tmp/mydir
/cvmfs/oasis.opensciencegrid.org/mis/apptainer/bin/apptainer build⮐
--sandbox ./root docker://cern/cc7-base
- The Apptainer User Guide
- https://apptainer.org/docs/user/main/index.html
Data management
Redundancy
- Information, research results, work, etc. should go onto persistent media.
- Beware: all devices will break at some point. Your personal devices get lost, stolen or have coffee spilled over them.
- Data centre hard drives break down all the time too.
Other threats
Very real emerging threat of ransomware attacks, data gets encrypted and may not be recoverable.
Redundancy is key; with sufficient redundancy the risk of data loss can be brought down to a minimum.
- data duplication across systems
- in geographically separated areas (to mitigate natural disasters)
- separated by organisational domains to mitigate ransomware attacks
Storage classes
From closest to furthest, we have
| location | bytes | Nikhef | stoomboot | external | backup |
|---|---|---|---|---|---|
| /home | \(O(10^{10})\) | ✓ | ✓ | ✓ | |
| /project | \(O(10^{12})\) | ✓ | ✓ | ✓ | |
| /data | \(O(10^{13})\) | ✓ | ✓ | ||
| /dcache | \(O(10^{14})\) | ✓ | ✓* |
(* you need to request external access)
Data access
Home, project and data directories are available via NFS, or network file system, only.
pros: simply access as a file system
cons: no access other than by file system, no high throughput scale
dCache
Dcache is a distributed storage system. It is therefore highly suited for high-throughput use and scales well with use in combination with the stoomboot batch system.
dCache
DCache is available through several protocols.
Via NFS under /dcache, but: not a pure POSIX file system. You cannot open a file for re-writing, i.e. no
f = open("/dcache/myproject/mydata/file.dat", O_RDWR); /* NO! */
but you can overwrite a file:
f = open("/dcache/myproject/mydata/file.dat", O_RDWR|O_TRUNC); /* ok */
dCache
Other access methods are available, most notably:
XRootD
xrdcp ./myfile.dat xroot://stam.nikhef.nl:/dcache/myproject/mydata/
WebDAV
davix-ls --cert $X509_USER_PROXY https://stam.nikhef.nl:2880/dcache
Both require the use of a grid certificate or grid proxy.
dCache remote access
(Subject to change; we may automate this in the near future!)
By request:
- controlled via grid certificate
- certificate subject name gets mapped to your nikhef user account
- send access request and your subject name to stbc-admin@nikhef.nl
Pitfalls

Running multiple threads on a single core job.
Some software is not multi-user aware. When it is run on a machine with 64 cores, it assumes all cores are yours to use. It will then go on to fire of as many threads, but since the job is pinned to a single core, all threads are now in contention for cycles on that core.
Check if the software has a setting to control the number of threads, or run jobs on whole nodes by specifying you need all the cores:
qsub -l nodes=1:ppn=32 job.sh
Be sure to match the ppn option to the actual number of cores on the machine.
We have 32 and 64 core systems, the latter served by the smefit queue.
Scaling up without knowing where the next choke point is
Doing things 'at scale' brings interesting challenges and problems, probably some of which you have never considered before.
Don't assume things will just work after a few small test.
The only way to learn what they are is to ramp up gradually to the point where things start to break down. At that point, investigate the issue and implement a mitigation before scaling up further.
Although the sysadmins are always curious to know what you are doing, we do not necessarily become involved in your scalability problems. Since our main concern is with the availability of the systems to all our users, our interventions may be swift and brutal.
Forgetting to turn off debugging…
…and flooding stdout/stderr
It is actually quite common that some code path in the innermost loop of a job prints out a useless message to stdout, resulting in a 100Hz flood of bytes to a file that was never meant to be that large. After it fills up its corner of the file system, the node and all its jobs grind to a halt until the sysadmin intervenes.
The stdout/stderr streams are actually files that are written
back to the submit host at the end of the job and as a rule of
thumb should remain very small (less than 1MB). There are many
options for data storage elsewhere (/data, /project,
/dcache, …)
Using /data and /project at scale
These servers are not distributed and they cannot handle hundreds and thousands of jobs.
Use the job's own local directory on the worker node; there are several TB of space available shared for all the job slots.
This data gets wiped at the end of a job.
Extremely short jobs
How large should a batch job be?
- Not too long, because nodes need to be cleared for maintenance at regular intervals (security updates)
- certainly not too short; there is \(O(\text{minutes})\) of overhead for a job, and a flood of extremely short jobs can overwhelm our aged batch system
At the minimum (for production runs) a job should last 10 minutes. Ideally, anything between 1 and 10 hours is perfect.
Beyond stoomboot
Would you believe there is actually an even bigger compute cluster than stoomboot available? Approximately 4 times the size?
We have this as part of the NL-T1 compute facility for Atlas, LHCb and Alice combined with the Dutch National Infrastructure. Collectively called 'the Grid'.
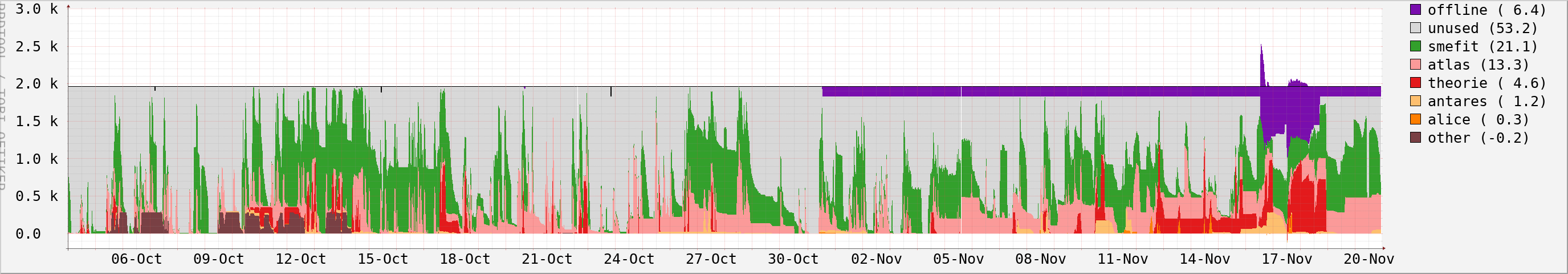
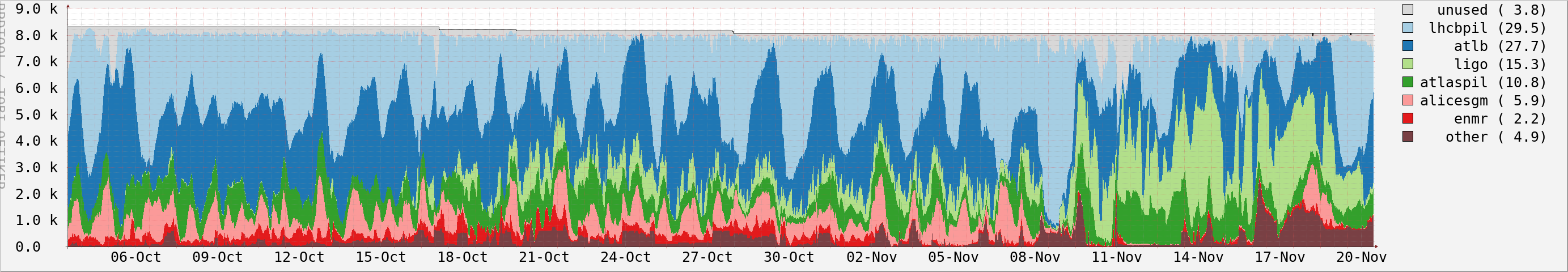
Stoomboot vs Grid
Stoomboot:
Grid:
Grid access
Some (most) of the Nikhef collaborations are eligible to use it (not only at Nikhef, but in other places as well!) so this could be a massive accelerator for your science.
Grid considerations
There are some caveats:
- no access to /home, /project, or /data
- access to dCache only through WebDAV/XRootD
- running jobs requires another interface through ARC, DIRAC, or other (experiment-specific) frameworks
Consider whether this might be worth your investment.
Fin
(we've made it)